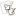RFLP Analysis of the cox1 and cox2 Gene Cluster
This procedure works well for identification of isolates to a species level when there are pure cultures to extract the DNA from. The current amplification primers will amplify plant sequences as well, so it should not be used with DNA extracted from plant samples. Sequence alignments suggest that it would be possible to develop primers that would not amplify plant sequences, but due to the low level of pathogen target DNA concentrations found in many field samples it will likely require that two rounds of amplification be done to generate enough DNA to use for RFLP analysis.
Amplification Primers
- Forward primer - FM 75 (dCCTTGGCAATTAGGATTTCAAGAT)
- Located in the 5' end of the cox II gene (starting at base 55 of the gene in P. infestans, NC002387)
- Reverse primers -.FM 77 (d CACCAATAAAGAATAACCAAAAATG) and FM 83 (dCTCCAATAAAAAATAACCAAAAATG)
- Located in the 3' end of the cox I gene (starting at base 1261 of the gene in P. infestans, NC002387)
- While FM 75 and FM 77 amplify most species, P. capsici, P. cinnamomi, P. citricola, and P. colocasiae were amplified better using FM 83 instead of FM 77
- A mixture of both primers is generally used in the amplification.
- Amplicon is approximately 2.2 kb in size depending on the species
- the spacer between the cox I and II genes can be variable in length among species
- Amplifies best with 3 mM MgCl2 and annealing temperature of 60ºC
- Amplification reactions were done in 50 µl and contained approximately 20 to50 ng of DNA, a final concentration of 1 µM of the forward and reverse primers, 5 µl of 10X buffer, 100 µM of each dNTP, 3 mM MgCl2, and 3 units of Taq DNA polymerase.
- Amplifications were done with a 3°C/s ramping time using the following parameters: one cycle at 95º C for 3 min; 35 cycles of 1 min annealing at 60ºC, 1 min extension at 72º C, and 1 min denaturation at 94ºC; followed by one extension cycle at 72ºC for 10 min.
- More recent work has identified isolates that do not amplify well under these conditions. While reducing the annealing temperature can alleviate this problem for some isolates, new primers are under evaluation to broaden the number of species that will be amplified and will be posted when the work has been completed.
- 4 µl of the amplification reaction was run on a 1.5% agarose gel to confirm the quality of the amplification and quantification of the amplicon.
- These primers do not amplify bands from a range of different plant species, but they do amplify bands from the related genus Pythium
Restriction Analysis
- Digestions with restriction
enzymes were conducted overnight in a total volume of 17 µl in accordance
with the manufacturer’s recommendations (New England Biolabs, Beverly, MA)
- Digested fragments were
separated in 3% NuSieve 3:1 agarose in 0.5X TBE buffer at 45 V for 6 h or
until the bromophenol blue dye in the loading buffer had migrated 8 cm
from the well.
- A 100-bp ladder mixed
with a nondigested amplicon from P. infestans (isolate 580;
approximately 2.2 kb based on sequence analysis) was used as size markers.
- The gel was stained in
ethidium bromide (0.5 µg/ml) for 30 min, destained in deionized water for
30 min, and photographed under short wave UV using either Polaroid Type 55
film or a digital imaging system .
- Scanned digital images of
the Polaroid negatives or the digital images were imported into the
computer program BioNumerics (ver. 2.5, Applied Maths, Sint-Martens-Latem,
Belgium). This computer program automatically determines the
molecular size of the RFLP bands relative to molecular size standards
included on each agarose gel.
- Bands less than 100
bp in size were excluded from the analysis due to the diffuse nature of
the bands when using a 3% NuSieve 3:1 agarose gel.
- Using a 5% gel these
small fragments can be clearly separated and can increase the
discrimination among species
- The images were processed
using standard procedures and molecular size determinations of digested
bands done automatically with manual confirmation.
- To optimize band
matching between isolates, the positional tolerance of each band (the
maximum shift between two bands that is allowed to consider the bands
matching, expressed as a percentage of the length of the gel run) and
optimization of the data (this allows for a shift between any two
patterns to optimize comparisons of banding patterns, expressed as a
percentage of the run length) was determined from the data using the
tolerance and optimization options of the program.
- Positional
tolerances of 1.06%, 1.0%, 1.16%, and 1.0% were obtained for AluI,
MspI, RsaI, and TaqI, respectively.
- Optimization was
determined to be 0 except for RsaI, which was set at 0.54%.
Results
- Digests with AluI alone could generate a species-specific diagnostic banding profiles able to differentiate most species evaluated in this investigation (see table below).
- A total of four restriction enzymes were used to increase the level of variation observed among species and improve the resolution of the technique (Fig. 2 for cluster analysis).
![]()
![]()
Digestion with AluI
|
Phytophthora sp. |
Isolate # |
AluI |
MspI |
RsaI |
TaqI |
|
P. arecae |
441 |
648, 460, 238, 227, 203, 143, 117 |
2190 |
663, 550, 332, 300, 226, 182 |
2184 |
| P. boehmeriae |
325 |
565, 355, 315, 238, 227, 201, 179, 164 |
1305, 1016 |
621, 550, 332, 290, 182 |
1870, 437 |
|
P. cactorum |
311 |
944, 327, 227, 201, 153, 132 |
1145 |
684, 550, 332, 300, 182, 148 |
2184 |
|
|
385 |
944, 327, 227, 201, 153, 132 |
1145, 564 |
684, 550, 332, 300, 182, 148 |
2184 |
|
P. capsici |
302 |
697, 451, 227, 203, 179, 132 |
2190 |
712, 550, 332, 300, 226, 182 |
1573, 743 |
|
|
307 |
697, 451, 227, 203, 179, 132 |
1415, 901 |
712, 550, 332, 300, 226, 182 |
1573, 743 |
|
|
Cp-1 |
697, 451, 227, 203, 179, 132 |
1140, 1080 |
712, 550, 332, 300, 226, 182 |
1187, 834 |
| P. cinnamomi |
448 |
697, 421, 267, 202, 179, 132 |
1145, 755, 412 |
712, 586, 405, 332, 300 |
2004, 319 |
|
P. citricola |
SB2078 |
697, 267, 227, 203, 179, 132 |
2190 |
783, 586, 332, 300, 182, 148 |
2184 |
|
|
Cr-4 |
697, 289, 267, 227, 203, 179, 132 |
2190 |
783, 586, 332, 300, 182, 148 |
2184 |
|
P. citrophthora |
414 |
697, 267, 227, 203, 179, 132 |
2190 |
712, 550, 332, 300, 226, 182 |
2184 |
| P. colocasiae |
350 |
565, 354, 315, 203 |
1533, 532, 218 |
712, 550, 332, 290, 226, 182 |
1870, 457 |
|
P. cryptogea |
438 |
697, 466, 298, 227, 179 |
1679, 564 |
712, 550, 332, 300, 226, 182 |
1619, 743 |
|
P. drechsleri |
439 |
697, 298, 227, 201, 153, 143 |
1333, 564, 364 |
712, 550, 332, 300, 226, 182 |
2004, 373 |
|
P. erythroseptica |
368 |
697, 466, 298, 227, 179 |
1679, 564 |
712, 550, 332, 300, 226, 182 |
1619, 743 |
|
|
365 |
697, 298, 267, 245, 227, 201 |
1679, 564 |
712, 550, 332, 300, 226, 182 |
1619, 743 |
|
P. fragariae var. fragariae |
393 |
665, 354, 327, 227, 203, 179, 153, 117 |
1145, 673, 490 |
712, 332, 290, 262, 226, 178, 125 |
2184 |
|
P. fragariae
var. rubi |
397 |
665, 421, 354, 227, 203, 179, 153, 117 |
1533, 603, 155 |
684, 332, 290, 262, 226, 178, 125 |
2184 |
|
P. gonapodyides |
393 |
697, 466, 227, 203, 153 |
2101, 211 |
684, 550, 332, 300, 116 |
1278, 591, 437 |
|
P. heveae |
Hv-2 |
665, 466, 298, 227, 179, 132, 117 |
2190 |
712, 550, 332, 300, 226, 182 |
1801, 284, 205 |
| P. hibernalis |
378 |
697, 466, 298, 227, 143 |
1533, 445, 327 |
621, 550, 332, 299, 182, 148 |
1120, 825, 373 |
| P. ilicis |
353 |
851, 354, 327, 227, 201, 143 |
1679, 673 |
663, 550, 332, 300, 182, 153 |
2004, 278 |
|
P. infestans |
550 |
944, 327, 187, 153 |
2190 |
684, 351, 332, 300, 243, 226, 178 |
1503, 591, 205 |
|
P. lateralis |
440 |
666, 267, 201, 179, 143, 117 |
1533, 755 |
621, 550, 332, 170, 134, 125 |
914, 743, 373, 205 |
|
P. megasperma |
336 |
697, 298, 267, 227, 201, 164, 143 |
1969, 211, 122 |
712, 550, 332, 300, 226, 182 |
2184 |
|
|
309 |
697, 466, 354, 179, 143 |
1016, 564, 445, 327 |
684, 550, 332, 300, 182, 153 |
1870, 392 |
|
|
437 |
697, 466, 370, 164 |
1016, 755, 532 |
823, 550, 332, 300, 182 |
1278, 591, 392 |
|
|
335 |
697, 451, 298, 250, 209, 179, 132 |
1533, 532, 211 |
663, 550, 332, 300, 182 |
1870, 437 |
|
P. megakarya |
327 |
697, 665, 209, 153 |
1071, 673, 564 |
663, 550, 405, 332, 300, 148 |
2004, 548, 205 |
|
|
328 |
648, 327, 201, 187, 153 |
1785, 532 |
663, 550, 332, 300, 226, 182 |
2004, 550, 205 |
|
P. mirabilis |
354 |
944, 421, 201, 153 |
2190 |
684, 351, 332, 300, 226, 182 |
1703, 591 |
|
P. nemorosa |
P-13 |
962, 356, 229, 202, 178, 149 |
2190 |
648, 553, 332, 226, 181 |
2184 |
|
P. nicotianae |
361 |
648, 466, 315, 297, 203, 179, 153 |
2190 |
684, 550, 332, 226, 170, 125 |
2184 |
|
P. palmivora |
329 |
648, 466, 238, 227, 201, 143, 117 |
1679, 564 |
663, 550, 332, 300, 226, 182 |
2184 |
|
|
Pl-14 |
648, 466, 238, 227,201, 143, 117 |
2190 |
663, 550, 332, 300, 226, 182 |
2184 |
|
P. phaseoli |
373 |
944, 327, 315, 201 |
1305, 532, 490 |
684, 351, 332, 300, 243, 226, 182 |
1703, 591 |
|
P. pseudosyringae |
470 |
944, 327, 250, 238, 209, 179 |
2190 |
823, 550, 332, 300, 182 |
2184 |
|
P. pseudotsugae |
308 |
944, 354, 227, 201, 132 |
2190 |
684, 550, 332, 300, 226, 182 |
2184 |
|
P. sojae |
312 |
697, 451, 298, 250, 203, 179, 132 |
2190 |
684, 550, 406, 332, 300 |
2184 |
|
P. ramorum |
|
697, 466, 298, 187, 132 |
1305, 954 |
621, 550, 332, 226, 178, 125 |
2184 |
|
P. syringae |
442 |
852, 315, 267, 227, 203 |
2190 |
1101, 550, 332, 300 |
1187, 1120 |
|
|
468 |
852, 315, 267, 227, 210, 152 |
2190 |
1101, 550, 332, 300 |
1187, 1019, 113 |
|
|
469 |
944, 327, 250, 209, 179 |
2190 |
762, 550, 332, 300 |
1187, 1120 |
a Amplified products were digested with the indicated restriction enzyme and separated in a 3% NuSieve 3:1 agarose gel. Fragment sizes were determined and the database managed using the computer program BioNumerics (ver. 2.5). While fragment sizes are reported to the base pair by BioNumerics, this level of accuracy is artificial and not supported by the agarose electrophoresis method used for estimation. Doublet bands and fragment sizes smaller than 100 bp are not reported or included in the cluster analysis.
|